One of the greatest paradigm shifts in medicine over the past few decades has been the unfolding discoveries revealing the metabolic influence of the human microbiome, especially that which resides within the gastrointestinal tract. Indeed, it is difficult to find a medical discipline that is not actively investigating the potential role played by the gut microbiome in human health and disease. This explosion of knowledge has been welcome news for many healthcare providers, though keeping up with the published research, changing nomenclature and therapeutic ramification of this information has been a difficult task. Along with this new fascination for all things microbiome related comes the natural inclination to “assess” and “fix” this complex ecosystem as one might do with other systems of the body. Here is where we need to slow down and ask whether we know enough about what is “normal,” what is “fixable,” and what may just be an adaptation to a person’s lifestyle inputs.
Most integrative and functional medicine clinicians are familiar with the term dysbiosis. Essentially, it describes any significant imbalance in the gut microbial ecosystem, especially one that leads to a negative host response. This includes either overgrowth or depletion of a particular commensal species, family or phylum of bacteria, or a geographic dislocation of one or more species (i.e., colon bacteria colonizing the small intestines). While an infection by a pathogenic microbe such as Salmonella is not usually called “dysbiosis,” the opportunistic overgrowth of bacteria like Clostridium difficile or a yeast like Candida albicans is often directly related to an infection or an antibiotic-induced alteration in the gut microbiota (i.e., dysbiosis-induced). Ironically, with the advanced technologies now available to help the clinician analyze specific changes in the gut microbiota, this term has almost become too generic for the research setting, and clinicians should be aware that more specific terms might be used to define specific microbiome-host dysfunctions. Still, the notion that a disturbance in the gut microbial ecosystem—dysbiosis— may be a major trigger in a wide range of gastrointestinal and systemic disorders is an important factor often missed by clinicians uninformed by these recent discoveries.
However, as the technology used to analyze an individual’s gut microbiota advances, we are finding more and more subtle differences in the amounts and ratios of species; some of which are associated with various chronic disease conditions. Properly understood, many of these subtle changes are microbial adaptations to the person’s lifestyle and metabolic signals; though others might deem them “dysbiosis.” Here are a few things to consider when trying to understand the stability, adaptability and the alterability of the human gut microbiome.
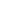
Before discussing the many factors that can alter the gut microbiome, it is important to establish the relative stability of a person’s microbiota. This is especially important for the clinician using fecal samples to analyze a person’s microbiota, and whether this analysis represents only a transient snapshot or are semi-permanent features of the patient’s core microbiome. Recently, several studies have attempted to answer this question by analyzing the microbiome (via fecal samples) of the same individuals over time, often tracking dietary or other changes along the way. In general, they have discovered that a fairly large core microbiome (~40 species of bacteria) remains stable in an individual over at least one year. In fact, a landmark study published in Science (2013) using fecal samples from 37 individuals showed that a person’s “core” gut microbiota is remarkably stable for greater than five years (they speculate for decades). They also showed that the core species found in close family members is very similar, and that species within the Bacteroidetes and Actinobacteria phyla are more stable than those in other phyla (e.g., Firmicutes).
One of the most comprehensive analyses of the core microbiome of adults was published recently by Falony et al. called the Flemish Gut Flora Project (FGFP). The study was originally designed to investigate the global core microbiome and to study the impact of host and environmental factors on microbiota variation within an average, healthy Western European population in Flanders, Belgium. Of the 503 metadata variables that emerged, 69 factors were shown to correlate significantly with overall microbiome community variation. The association between BMI and microbiome composition was found to be small but significant in effect size. Of all the covariates detected, 63% were driven by medication (mostly the use of antibiotics and laxatives), making the use of any medication the largest predefined explanatory variable tested in this study. However, when breaking down the covariates into smaller categories, stool consistency as measured by the Bristol Stool Scale (BSS) emerged as the top single non-redundant microbiome covariate in the FGFP metadata. BSS score reflects transit time, water availability, and potential niche differentiation within the colon ecosystem. For 12 of the 20 FGFP core genera, core abundance was shown to increase with looser stools. The authors note medication use and BSS scores have been largely ignored in microbiome studies, and should be considered in future studies based on the strong findings in the FGFP cohort.
The connection between stool consistency and microbiota abundance/diversity has been further analyzed. Vandeputte et al. (2016) found stool consistency (as measured by BSS) was strongly associated with fecal microbial richness. Species richness was shown to significantly decline as stool firmness declined (p=0.0007), reaching its minimum in those with diarrhea. The group also found enterotypes were distributed over BSS scores, the Prevotella enterotype was more abundant in looser stools (p=0.019) and the Ruminococcaceae-Bacteroides (RB) enterotype completely dominated firmer samples (p=0.019) perhaps suggesting that enterotypes may be a surrogate marker for bowel transit time since transit time may select for bacteria with certain traits. For instance, individuals with a short transit time may have greater amounts of fast-growing bacterial species, while those with slow transit times may instead select bacteria with greater adherence to host tissue.
What this and other data seem to reveal is that while a core microbiome (structure and species) is part of development, and is formed as part of an individual’s early inoculation and environmental exposure, subtle and important changes in the variable commensal microbiome can occur without fundamentally altering the long-term stability of the core species (at least as determined by analysis of fecal microbiota). This means differences in fecal microbiota (beyond the core species enumeration and ratio) may reflect adaptations to environmental or pathophysiological changes that can be meaningfully followed in clinical research or in clinical application. Lastly, there is also a significant portion of the microbiota that is transient; most comes from our food (including probiotics), water, air, and environment. These microorganisms rarely persist beyond a week or two, but they can be important modulators of both the core and variable microbiome, and greatly influence host physiology.
Therefore, sampling and analyzing fecal microbiota as a routine part of a yearly physical exam may be a way to note subtle (or radical) changes in the microbiome consistent with noted changes in a patient’s overall health. We predict microbiome testing will soon be a routine practice for annual exams, like standard blood tests, and will be part of the health assessment of most subjects.
This is a very tricky question. If the question implies a basic set of microbial species that is ideal for any human subject living anywhere, then the answer is “no.” However, we believe there is likely a suitable microbiome for each person, one that is properly adapted to their particular geography, genetics, diet and environment. In fact, if we are correct in understanding the microbiome as the ultimate adaptable organ system, the microbiota within the GI tract (and therefore in the fecal sample) should change with age, with seasonal dietary changes and with health status. Comparing the microbiome of Western subjects consuming monotonous and poor diets to the microbiome of hunter-gatherers eating seasonally shows dramatic differences. While some would describe the microbiota of these hunter-gatherers as ideal, it probably would be poorly sustained and suited to benefit the Western subject eating a standard Western diet.
As we shall see, general changes in the fecal microbiota are indicative of certain gastrointestinal disorders (when compared to “healthy” control individuals). Some of these alterations reflect wholesale changes at the phylum-level, while others are subtle changes in one or two species. Most of the time, however, the first analysis of a patient’s microbiome is performed after a dysfunctional change has already occurred (often the first encounter with the patient) and therefore clinicians do not usually have access to a patient’s former (“baseline”) core microbiome for comparison. Nonetheless, analyzing gut microbiota (via fecal microbiota testing) is often helpful in both diagnosing a complex GI-related issue, as well as for confirming therapeutic impact (via follow-up testing).
When an imbalance in the number and types of commensal microbes develops in the gut, the immune system is activated to respond in a variety of ways. New research now suggests that in some patients, the immune system can misinterpret a “normal” microbiome as “dysbiotic” due to genetic polymorphisms in a variety of immune receptors tasked with interpreting the gut microbial environment. Pattern recognition receptors are an important component of both the innate and adaptive immune response, and are an important mechanism for helping the host immune system interface with the gut microbiome. However, similar to an overreaction to a benign food protein that causes an immune response in some individuals (based on genetics), certain commensal organisms may trigger an inappropriate response when the host immune system misinterprets their molecular pattern as “pathogenic” (e.g., inflammation, auto-immune cross-reactivity, etc.).
Likewise, inappropriate receptor expression caused by changes in genetics (gene/protein sequence) or genomics (altered expression pattern of one or more receptor) can result in dysfunctional immune regulation of the commensal organisms within the gut (e.g., reduced levels of sIgA to target pathobionts). This lowers the precision of the immune surveillance within the GI tract, allowing for an imbalance in the commensal organisms; creating a host-derived genetics-induced dysbiosis. Clinicians attempting to help correct a patient’s gut dysbiosis must understand that host factors, including genetics, may profoundly affect how they respond to therapies intended to re-balance the gut microbiome (via diet, probiotics, prebiotics, etc.). Therefore, some recommendations or products may work better or quicker (or not at all) in some patients as compared to others.
This post is a modified excerpt of Dr. Guilliams’ new book Functional Strategies for the Management of Gastrointestinal Disorders, designed to help clinicians navigate the principles and protocols of the functional and metabolic medicine approach to the complex topic of GI health.
- 1Martínez I, Muller CE, Walter J. Long-term temporal analysis of the human fecal microbiota revealed a stable core of dominant bacterial species. PLoS One. 2013 Jul 16;8(7):e69621.
- 2Faith JJ, Guruge JL, Charbonneau M, et al. The long-term stability of the human gut microbiota. Science. 2013 Jul 5;341(6141):1237439.
- 3Falony G, Joossens M, Vieira-Silva S, et al. Population-level analysis of gut microbiome variation. Science. 2016 Apr 29;352(6285):560-4.
- 4Vandeputte D, Falony G, Vieira-Silva S, et al. Stool consistency is strongly associated with gut microbiota richness and composition, enterotypes and bacterial growth rates. Gut. 2016 Jan;65(1):57-62.
- 5Nibali L, Henderson B, Sadiq ST, Donos N. Genetic dysbiosis: the role of microbial insults in chronic inflammatory diseases. J Oral Microbiol. 2014 Feb 25;6.
- 6Frosali S, Pagliari D, Gambassi G, et al. How the Intricate Interaction among Toll-Like Receptors, Microbiota, and Intestinal Immunity Can Influence Gastrointestinal Pathology. J Immunol Res. 2015;2015:489821.
 | About Thomas G. Guilliams Ph.D. Dr. Guilliams earned his doctorate from the Medical College of Wisconsin (Milwaukee) where he studied molecular immunology in the Microbiology Department. Since 1996, he has spent his time studying the mechanisms and actions of natural-based therapies, and is an expert in the therapeutic uses of nutritional supplements. As the Vice President of Scientific Affairs for Ortho Molecular Products, he has developed a wide array of products and programs which allow clinicians to use nutritional supplements and lifestyle interventions as safe, evidence-based and effective tools for a variety of patients. Tom teaches at the University of Wisconsin-School of Pharmacy, where he holds an appointment as a Clinical Instructor; at the University of Minnesota School of Pharmacy and is a faculty member of the Fellowship in Anti-aging Regenerative and Functional Medicine. He lives outside of Stevens Point, Wisconsin with his wife and children. | |